Generators Interface¶
- class RandomEngine(seed: int = None)¶
Mersenne Twister pseudo-random generator of 32-bit numbers with a state size of 19937 bits.
- Parameters
seed – Random number generator seed; defaults to a call to random_device.
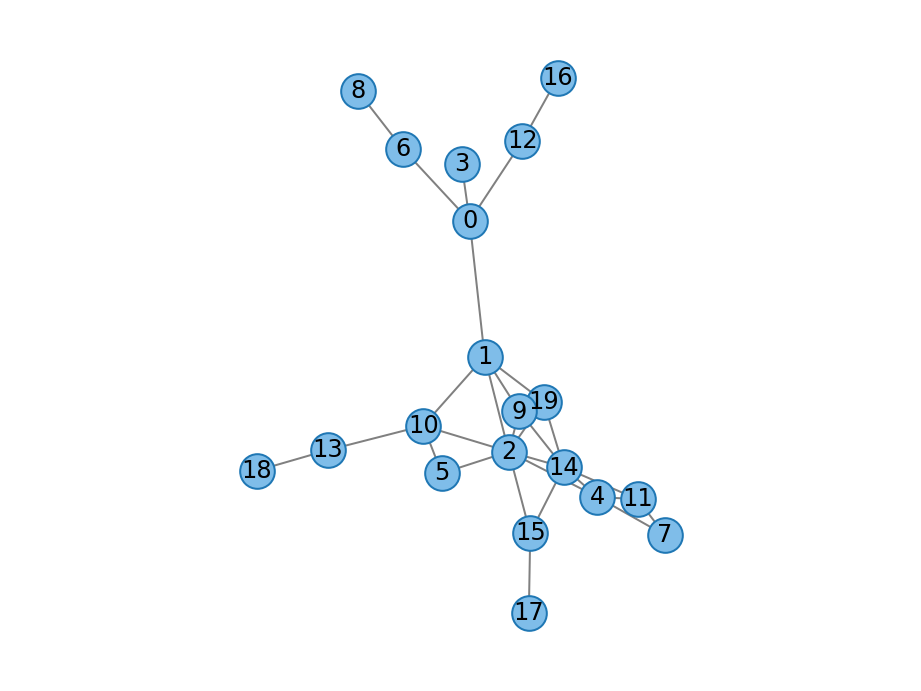
- duplication_complementation_graph(count_t n: count_t, double deletion_proba: float, double interaction_proba: float, Graph graph: Graph = None, random_engine=None) Graph¶
” Duplication divergence graph with complementation as described by [Vazquez2003].
- Parameters
n – Number of nodes.
deletion_proba – Probability that a duplicated or original edge is deleted (\(q\) in [Vazquez2003]).
interaction_proba – Probability that the original and duplicated node are connected (\(p\) in [Vazquez2003])
graph – Seed graph; defaults to a pair of connected nodes.
random_engine – See
get_random_engine().
The growth process proceeds in four stages at each step \(t\):
A node \(i\) is chosen at random and duplicated to obtain a new node \(t\), including its connections.
For each neighbors \(j\), we choose one of the edges \((i, j)\) and \((t, j)\) and remove it with probability deletion_proba. I.e., we remove at most one of the edges.
The nodes \(i\) and \(t\) are connected with probability interaction_proba.
Discard the new node \(t\) if it does not have any edges. This makes it more likely but cannot guarantee that the graph is connected. The strategy is adpated from [Ispolatov2005] who considered a simpler model without edge deletion. See
duplication_mutation_graph()for further details.
- Ispolatov2005
I. Ispolatov, P. L. Krapivsky, and A. Yuryev. Duplication-divergence model of protein interaction network. Phys. Rev. E, 71(6):061911, 2005. https://doi.org/10.1103/PhysRevE.71.061911
- Vazquez2003(1,2,3)
A. Vazquez, A. Flammini, A. Maritan, and A. Vespignani. Modeling of protein interaction networks. Complexus, 1(1):38–44, 2003. https://doi.org/10.1159/000067642
(Source code, png)
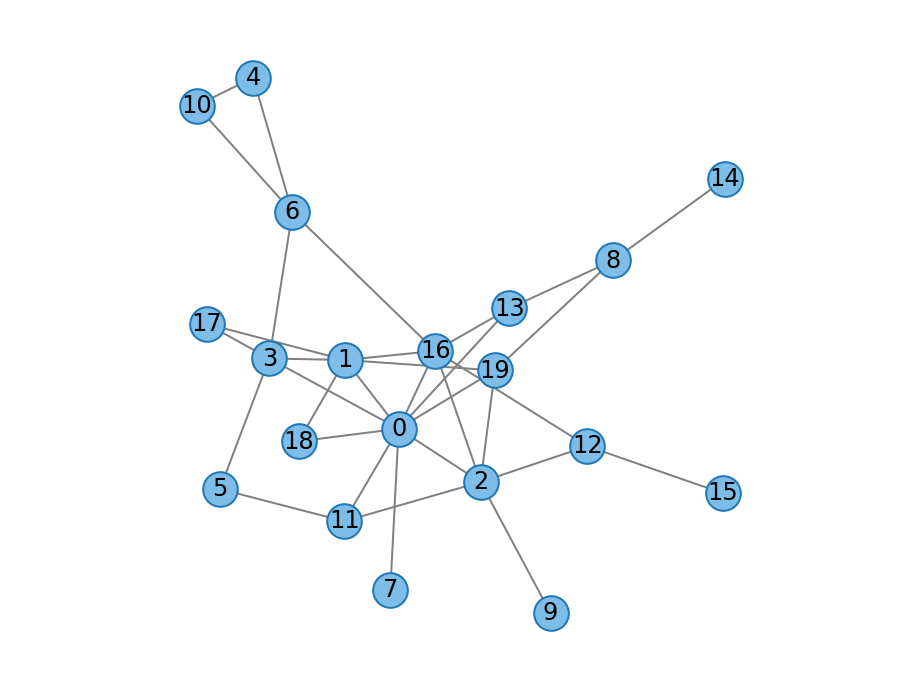
- duplication_mutation_graph(n: int, double deletion_proba: float, double mutation_proba: float = 0, Graph graph: Graph = None, random_engine=None, drop_isolates: bool = False) Graph¶
Duplication divergence graph with random mutations as described by [Sole2002].
- Parameters
n – Number of nodes.
deletion_proba – Probability that a duplicated edge is deleted (\(\delta\) in [Sole2002]).
mutation_proba – Scaled mutation probability (\(\beta\) in [Sole2002]) such that connections between the new node and existings nodes are created with probability \(\min\left(1, \beta / t\right)\).
graph – Seed graph; defaults to a pair of connected nodes.
random_engine – See
get_random`_engine().drop_isolates – Whether to drop new nodes if they do not have any connections.
- Returns
graph – Graph generated by the duplication divergence model.
The growth process proceeds in four stages at each step \(t\):
A node \(i\) is chosen at random and duplicated to obtain a new node \(t\).
For each neighbor \(j\) of \(i\), a connection to the new node \(t\) is added with probability \(1 - \delta\).
Connections between the new node \(t\) and any other nodes in the network are created with probability \(\min\left(1, \beta / t\right)\).
Discard the new node \(t\) if it does not have any edges. This ensures the graph remains connected and is adapted from [Ispolatov2005]. Indeed, setting \(\beta = 0\) reduces to Ispolatov’s model and is equivalent to
networkx.generators.duplication.duplication_divergence_graph().
Note
In the third step, we sample the number of additional edges \(k\) from a binomial random variable with \(t - 1\) trials and probability \(\min\left(1, \beta / t\right)\). We then sample \(k\) neighbors with replacement and connect them to \(t\). The actual number of additional connections may thus be smaller than \(k\), especially when the graph is small. This compromise avoids relatively expensive sampling without replacement from the population of nodes.
- Ispolatov2005
I. Ispolatov, P. L. Krapivsky, and A. Yuryev. Duplication-divergence model of protein interaction network. Phys. Rev. E, 71(6):061911, 2005. https://doi.org/10.1103/PhysRevE.71.061911
- Sole2002(1,2,3)
R. V. Sol ́e, R. Pastor-Satorras, E. Smith, and T. B. Kepler. A model of large-scale proteome evolution. Adv. Complex Syst., 5(1):43–54, 2002. https://doi.org/10.1142/S021952590200047X
(Source code, png)
- get_random_engine(arg=None) RandomEngine¶
Utility function to get a random engine.
- Parameters
arg – One of None, an integer seed, or a
RandomEngine. If None, a default, global random engine is returned. If an integer seed, a newly seeded random engine is returned. If aRandomEngine, it is passed through.- Returns
random_engine – Initialized
RandomEngineinstance.
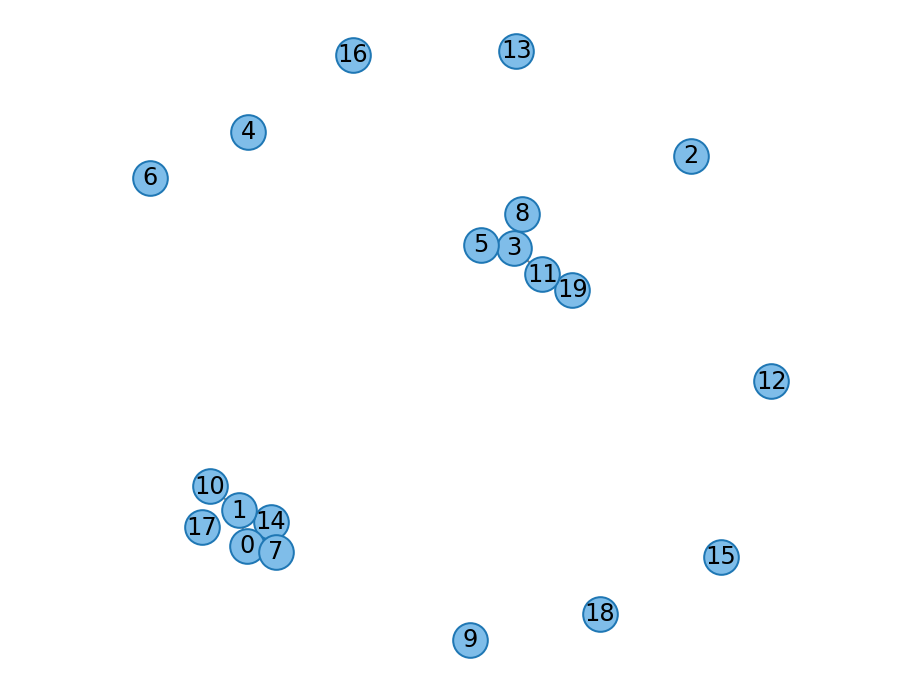
- gnp_random_graph(count_t n: count_t, double p: float, Graph graph: Graph = None, random_engine=None) Graph¶
Erdos-Renyi or \(G(n, p)\) graph. See
networkx.generators.random_graphs.gnp_random_graph()for details.- Parameters
n – Number of nodes.
p – Probability to create an edge between any pair nodes.
graph – Seed graph; defaults to the empty graph.
random_engine – See
get_random`_engine().
- Returns
graph – Graph generated by the \(G(n, p)\) model.
(Source code, png)
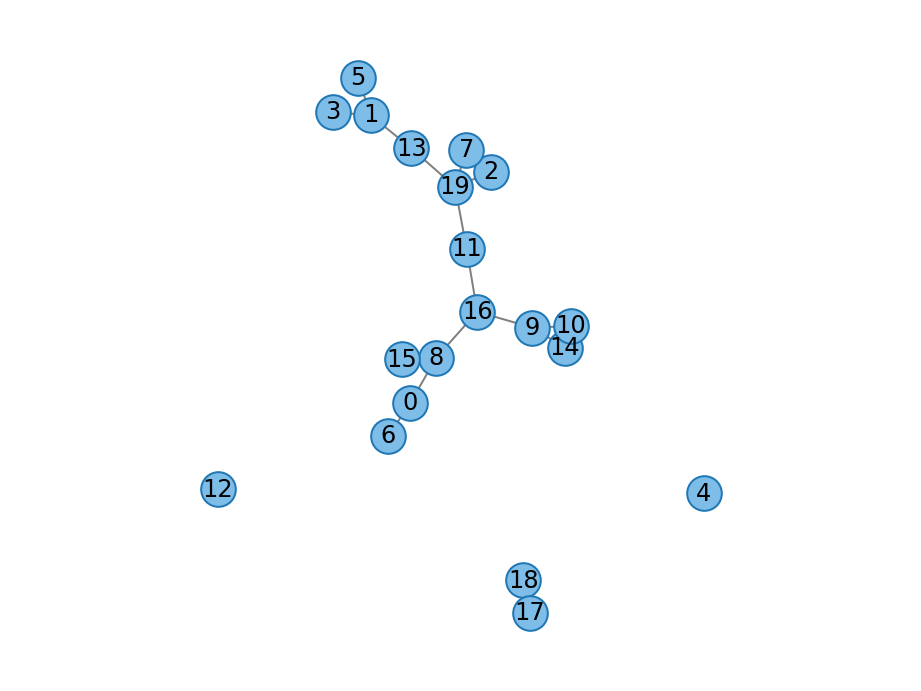
- redirection_graph(count_t n: count_t, double p: float, count_t m: count_t, Graph graph: Graph = None, random_engine=None) Graph¶
Redirection graph obtained by selecting random nodes and probabilistically redirecting to their neighbors before forming a connection.
- Parameters
n – Number of nodes.
p – Redirection probability.
m – Number of stubs for each new node.
graph – Seed graph; defaults to a graph with a single node.
random_engine – See
get_random`_engine().
- Returns
graph – Graph generated by the redirection model.
Note
For performance reasons, we sample both candidate nodes (before possible redirection) and nodes after redirection with replacement. The realized number of connections for a new node may thus be less than \(m\).
This generator is equivalent to the model proposed by [Krapivsky2001] implemented by
networkx.generators.random_graphs.gnr_graph()if \(m = 1\).- Krapivsky2001
P. L. Krapivsky and S. Redner. Organization of growing random networks. Phys. Rev. E, 63(6):066123, 2001. https://doi.org/10.1103/PhysRevE.63.066123
(Source code, png)
- surfer_graph(count_t num_nodes: count_t, double connection_proba: float, Graph graph: Graph = None, random_engine=None) Graph¶
Random surfer graph obtained by exploring the neighborhood of a seed node using a random walk and connecting to nodes in the process.
- Parameters
num_nodes – Number of nodes.
connection_proba – Probability of connecting to a candidate node at each step.
graph – Seed graph; defaults to a graph with a single node.
random_engine – See
get_random`_engine().
- Returns
graph – Graph generated by the surfer model.
This generator corresponds to model A of [Vazquez2003]. We do not implement the recursive search generator (model B) because of its computational complexity.
- Vazquez2003
A. Vazquez. Growing network with local rules: Preferential attachment, clustering hierarchy, and degree correlations. Phys. Rev. E, 67(5):056104, 2003. https://doi.org/10.1103/PhysRevE.67.056104
(Source code, png)